 Graduate Student: Cell Biology and Physiology (2017-present)
Graduate Student: Cell Biology and Physiology (2017-present)
Contact: ntanke22@live.unc.edu
Education Gettysburg College- Gettysburg, PA- B.S. in Biology, 2017
Research: My goal is to understand the mechanism of endothelial cell quiescence mediated by fluid shear stress. To do so, I am utilizing in vitro fluidic devices as well as in vivo zebrafish models. This work has important implications for understanding endothelial cell cycle specific changes during developmental angiogenesis and wound healing.
Publications:
Liu Z, Ruter DL, Quigley KM, Tanke N, Jiang Y, Bautch VL (2021). Single-Cell RNA Sequencing Reveals Endothelial Cell
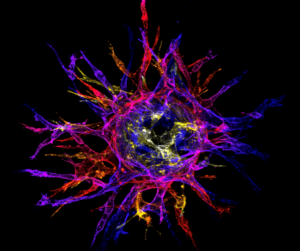
Transcriptome Heterogeneity under Homeostatic Laminar Flow. Arteriosclerosis, Thrombosis, and Vascular Biology. 2021 Aug 26:ATVBAHA121316797. Featured article of the issue.
Tanke, N. T.; Parfitt, G.; Umazume, T.; Segura, P.; Kalajzic, I.; Jester, J. V.; Dartt, D.; Makarenkova, H, P. “Unraveling lacrimal gland stem cell dynamics by lineage tracing.” 8th International Conference on the Tear Film & Ocular Surface,Montpellier, France, September 7-10, 2016. Abstract 74.
Delesalle, V.A., Tanke, N.T., Vill, A.C., and Krukonis, G.P. (2016). Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage 6, e1219441.
Pope, W.H., Bowman, C.A., Russell, D.A., Jacobs-Sera, D., Asai, D.J., Cresawn, S.G., Jr, W.R.J., Hendrix, R.W., Lawrence, J.G., Hatfull, G.F., et al. (2015). Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4, e06416.
Funding:
NRSA F31 Fellowship March 2021- present

